A cutting-edge Technology
The Science Behind FAST and splitfastOur technology is continuously fueled by the expertise detailed in our academic papers, published and discussed in international journals.
FAST and SPLITFAST AT A GLANCE
The Fluorescence-Activating and Absorption-Shifting Tag
FAST and splitFAST of The Twinkle Factory enable the specific fluorescent labeling of any protein of interest (POI). The protocol is based on the instantaneous formation of a fluorescent molecular assembly between the small (14 kDa), genetically encoded, protein tag FAST and various fluorogenic ligands (TFFluorogens).
TFFluorogens are dark in water and strongly fluoresce only when bound to FAST. They hence enable to detect and image FAST-tagged proteins with high contrast without the need of washing the excess of fluorogenic ligands. Furthermore, while labeling of FAST-tagged proteins with a TFFluorogen is non-covalent, it can easily be reversed by washing when necessary.
Furthermore, FAST does not require molecular oxygen for being fluorescent, unlike fluorescent proteins. FAST can hence fluorescence label proteins in weakly oxygenated or anaerobic environments. This is helpful for mature biofilm imaging, or for any anaerobe imaging, e.g., Clostridium.

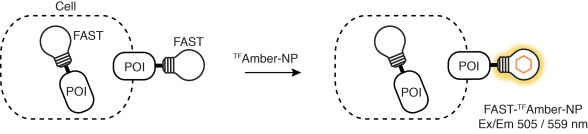
Furthermore, labeling of FAST-tagged proteins can be restricted to the cell surface excluding inner proteins. It indeed now exists TFFluorogens unable to cross the cell membrane, e.g., HBRAA-3E or tfAmber-NP. One can hence study protein trafficking at the membrane with various fluorimetric techniques, even including non-imaging flow cytometry.
Finally, on a practical standpoint, implementing FAST requires two steps:
1/ cloning and expressing the FAST-tagged protein, and
2/ labeling the resulting fusion with the TFFluorogen of choice.
Note that proteins of interest can be expressed with FAST as either an N- or a C-terminal fusion.
BEYOND FAST
splitFAST for protein-protein interactions
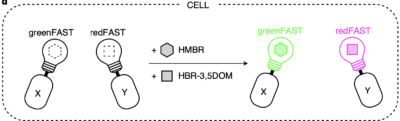
Prof. Arnaud Gautier reports greenFAST and redFAST, orthogonal variants of FAST for multicolor imaging, in Nature Chemical Biology. A powerful tool for cell division monitoring, sequential or multiple protein-protein interaction studies, high-content analysis, and much more!

Dr. Arnaud Gautier and Dr. Alison Tebo from ENS (Paris, France) report in Nature Commun. the development of splitFAST, a fluorescence complementation system for the visualization of transient protein-protein interactions in living cells. Engineered from the fluorogenic reporter FAST (Fluorescence-Activating and absorption-Shifting Tag), splitFAST displays rapid and reversible complementation, allowing the real-time visualization of both the formation and the dissociation of a protein assembly. The paper discloses various applications of splitFAST: interaction between a membrane protein and a cytosolic protein, interactions from the MAPK signaling pathway, real-time monitoring of the MEK1-ERK2 interaction, real-time monitoring of transient Ca-dependent interactions, apoptosis biosensor.
Beyond FRET and BiFC
Conventional imaging techniques for protein-protein interactions encompass Föster Resonance Energy Transfer (FRET) and bimolecular fluorescence complementation (BiFC), the latter being found easy to implement, straightforward to interpret, and less sensitive to the relative levels of the two interacting proteins. Now, in contrast to BiFC, splitFAST complementation was shown fully reversible and disassembly rapid, which allows not only the real-time monitoring of protein complex assembly but also the real-time monitoring of protein complex disassembly. This unprecedented behavior opens exciting prospects to study the role and function of protein-protein interactions in various cellular processes and dissect complex interaction networks.
splitFAST works with the classical fluorogens of The Twinkle Factory, reflecting the versatility of this breakthrough fluorescent reporting system.
ABOUT FAST VARIANTS
greenFAST and redFAST for multicolor orthogonal labeling
Prof. Arnaud Gautier reports greenFAST and redFAST, orthogonal variants of FAST for multicolor imaging, in Nature Chemical Biology. A powerful tool for cell division monitoring, sequential or multiple protein-protein interaction studies, high-content analysis, and much more!





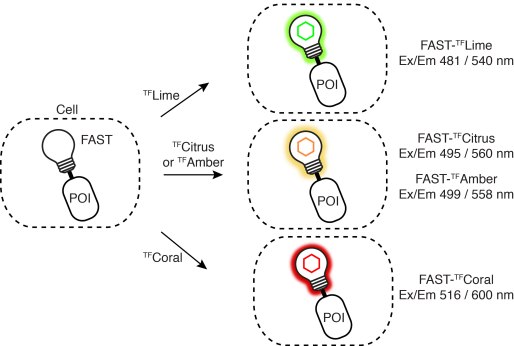
Driving collaborations with KU Leuven, Collège de France, University of Oxford, King’s College London, a team led by Dr. Alison Tebo (ENS, Paris) and Prof. Arnaud Gautier (Sorbonne University) was able to level up FAST into two orthogonal tags, greenFAST and redFAST. greenFAST recognizes specifically the fluorogen HMBR a.k.a. tfLime, thus forming a green fluorescent complex. And redFAST recognizes HBR-3,5DOM or tfCoral, forming an orange-red fluorescent complex. Moreover, their orthogonal absorption profiles also avoid any crosstalk in their one- or two-photon excitation.
The paper discloses various modalities of implementation of greenFAST & redFAST: two-color confocal microscopy, fluorescence lifetime imaging microscopy (FLIM), two-photon excitation, and even super-resolution microscopy (SOFI) with redFAST.
greenFAST / redFAST labeling seems particularly promising for dynamic recording in vivo. The authors apply it to the FUCCI (fluorescence ubiquitination cell cycle indicator) in mammalian cells and in zebrafish embryos. In the latter, it allowed exquisite monitoring of individual nuclei from the 256-cell stage through to the tenth cycle. In addition, fluorescence recording of the whole embryo revealed proliferation patterns and asynchronic division as early as 256 cells. Detailed analysis of cell cycles as short as 15 min and as early as 3.5 hpf is only possible with quasi-instantaneous fluorescence as provided by FAST and its variants.
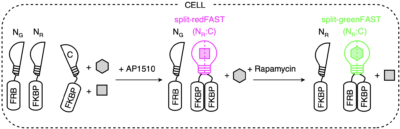
splitFAST, earlier published in Nature Communications, is the only reversible fluorescence complementation reporter with rapid association/dissociation kinetics. Aiming breakthrough biosensors capable to report sequential or multiple protein interactions, the authors have hence developed split tags, namely split-greenFAST and split-redFAST. To illustrate their potential, they have tracked the formation of FKBP-FKBP dimers mediated by AP1510. And further, their exchange to FRB-FKBP dimers upon the addition of rapamycin.
They have thus proven the ability of split-greenFAST / split-redFAST to deliver facile readout of multiple or sequential protein interactions with good contrast and temporal resolution.
ABOUT FAST VARIANTS – AGAIN
frFAST for far-red labeling
The latest FAST variant is frFAST designed together with the fluorogen HPAR-3OM, a.k.a. tfPoppy, for far-red labeling. Published by Prof. Arnaud Gautier in Angew. Chem. Int. Ed. 2020.





A team lead by Prof. Arnaud Gautier discloses frFAST for far-red labeling in Angew. Chem. Int. Ed. 2020. The team gathers researchers from Sorbonne University, and Collège de France and ENS of PSL Research University.
The high demand for RFP, and furthermore far-red fluorescent proteins, has emerged from the need for new colors. Indeed the growing use of optical biosensors and optogenetic tools obstruct the spectral window available for biological imaging. Moreover, far-red fluorescent proteins allow imaging deeper in tissues. Indeed, autofluorescence, light scattering and absorbance by endogenous molecules is reduced in the far-red region.
This paper discloses a far-red system, frFAST and HPAR-3OM, engineered from the original FAST tag and HBR-based fluorogens. HPAR-3OM is a membrane-permeant fluorogenic ligand which selectively binds frFAST-tagged proteins. It works in solution, in living cells or in fixed cells. HPAR-3OM is almost non-fluorescent when free in solution. But it strongly fluoresces when bound to frFAST. Gautier designed HPAR-3OM together with a variant of FAST, frFAST, specifically for far-red labeling. frFAST and HPAR-3OM allow for imaging proteins in mammalian cells, but also in zebrafish embryo/larvae and chicken embryo. Gautier eventually developed a rapid and reversible split-frFAST to image protein-protein interactions in live cells.
Wish to use frFAST or split-frFAST? The Twinkle Factory sells the fluorogen HPAR-3OM under the tradename tfPoppy, reference 570655-250.
The paper discloses various modalities of implementation of greenFAST & redFAST: two-color confocal microscopy, fluorescence lifetime imaging microscopy (FLIM), two-photon excitation, and even super-resolution microscopy (SOFI) with redFAST.
READ MORE
Peer-reviewed papers
Since the seminal PNAS in 2016 by the team of Arnaud Gautier, from the first enthusiastic mention in 2018 by Terry Papoutsakis of University of Delaware, navigate the fascinating journey of FAST adoption and buildup towards The Twinkle Factory community.





Papers by The Twinkle Factory team
- Nat. Commun. 2025 – El Hajji, L., Bunel, B., Joliot, O., Li, C., Tebo, A. G., Rampon, C., … & Gautier, A. A tunable and versatile chemogenetic near infrared fluorescent reporter.
- Adv. Sci. 2024 – El Hajji, L., Lam, F., Avtodeeva, M., Benaissa, H., Rampon, C., Volovitch, M., Vriz, S. and Gautier, A. Multiplexed in vivo imaging with Fluorescence Lifetime-Modulating tags.
- ACS Chem. Biol. 2024 – Rakotoarison, L. M., Tebo, A. G., Böken, D., Board, S., El Hajji, L., & Gautier, A. Improving Split Reporters of Protein–Protein Interactions through Orthology-Based Protein Engineering.
- bioRxiv 2023 –A tripartite chemogenetic fluorescent reporter for imaging ternary protein interactions
- ACS Sens. 2023 – Broch, F., El Hajji, L., Pietrancosta, N., & Gautier, A. Engineering of Tunable Allosteric-like Fluorogenic Protein Sensors
- Acc. Chem. Res. 2022 – Gautier, A. Fluorescence-Activating and Absorption-Shifting Tags for Advanced Imaging and Biosensing.
- Methods Mol. Biol. 2022 – in Yeast Surface Display – Hajji, L. E., Benaissa, H., & Gautier, A. Isolating and Engineering Fluorescence-Activating Proteins Using Yeast Surface Display.
- Nat. Commun. 2021 – Benaissa, H., Ounoughi, K., Aujard, I., Fischer, E., Goïame, R., Nguyen, J., … & Gautier, A. An engineered multifunctional protein tag for advanced fluorescence imaging.
- Angew. Chem. Int. Ed. 2020 – Li, C., Tebo, A. G., Thauvin, M., Plamont, M. A., Volovitch, M., Morin, X., … & Gautier, A. A far-red fluorescent chemogenetic reporter for in vivo molecular imaging.
- Nat. Chem. Biol. 2020 – Tebo, A. G., Moeyaert, B., Thauvin, M., Carlon-Andres, I., Bokën, D., Volovitch, M., … & Gautier, A. Orthogonal fluorescent chemogenetic reporters for multicolor imaging.
- ChemPlusChem 2020 – Broch, F., & Gautier, A. Illuminating cellular biochemistry: fluorogenic chemogenetic biosensors for biological imaging.
- Eur. J. Org. Chem. 2020 – Aissa, H. B., & Gautier, A. Engineering glowing chemogenetic hybrids for spying on cells.
- Nat. Commun. 2019 – Tebo, A. G., & Gautier, A. A split fluorescent reporter with rapid and reversible complementation.
- Bioconjug. Chem. 2018 – Li, C., Mourton, A., Plamont, M. A., Rodrigues, V., Aujard, I., Volovitch, M., … & Joliot, A. Fluorogenic Probing of Membrane Protein Trafficking.
- ACS Chem. Biol. 2018 – Tebo, A. G., Pimenta, F. M., Zoumpoulaki, M., Kikuti, C., Sirkia, H., Plamont, M. A., … & Gautier, A. Circularly permuted fluorogenic proteins for the design of modular biosensors.
- Biochemistry 2018 – Tebo, A. G., Pimenta, F. M., Zhang, Y., & Gautier, A. Improved chemical-genetic fluorescent markers for live cell microscopy.
- L’Act. Chim. 2018 – Gautier, A. Spying on cells with chemical-genetic hybrids (Espionner les cellules avec des hybrides chémogénétiques).
- Chem. Sci. 2017 – Li, C., Plamont, M. A., Sladitschek, H. L., Rodrigues, V., Aujard, I., Neveu, P., … & Gautier, A. Dynamic multicolor protein labeling in living cells.
- Proc. Natl. Acad. Sci. U.S.A. 2016 – Plamont, M. A., Billon-Denis, E., Maurin, S., Gauron, C., Pimenta, F. M., Specht, C. G., … & Moncoq, K. Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo.
Other research teams like FAST and splitFAST
- Russ. J. Bioorg. Chem. 2025 – Sokolinskaya, E. L., Bogdanova, Y. A., Myasnyanko, I. N., Sokolov, A. I., Krasnova, S. A., & Baranov, M. S. Nano-frFAST: Design of a New Genetically-Encoded Far-Red Fluorescent Label.
- Cell Syst. 2025 – Nevot, G., Santos-Moreno, J., Campamà-Sanz, N., Toloza, L., Parra-Cid, C., Jansen, P. A., … & Güell, M. Synthetically programmed antioxidant delivery by a domesticated skin commensal.
- chemRxiv 2025 – Kozma, E., Szatmári, Á., Novák, T., Török, G., Nikić-Spiegel, I., Kormos, A., … & Kele, P. Toward Far-red Emitting Chemogenetic Labelling for Live Cell Super-Resolution Microscopy using Fluorescence-Activating and Absorption-Shifting Tag.
- Appl. Microbiol. Biotech. 2025 – Schöllkopf, A. I., Almeida, L., Krammer, K., Rivero, C. G., Liebl, W., & Ehrenreich, A. Deletion of atypical type II restriction genes in Clostridium cellulovorans using a Cas9-based gene editing system.
- Research Square 2025 – Sitara, A., Hocq, R., Lu, A. J., & Pflügl, S. Hi-TARGET: A fast, efficient and versatile CRISPR type IB genome editing tool for the thermophilic acetogen Thermoanaerobacter kivui.
- bioRxiv 2024 – Kivimaki, S.E., Dempsey, S., Tani, J.M., Camper, C., Hicklin, I.K., Blaby-Haas, C.E., Brown, A.M. and Melville, S. Type IV pili-associated secretion of a biofilm matrix protein from Clostridium perfringens that forms intermolecular isopeptide bonds.
- Nat. Commun. 2024 – García Casas, P., Rossini, M., Påvénius, L., Saeed, M., Arnst, N., Sonda, S., Fernandes, T., D’Arsiè, I., Bruzzone, M., Berno, V. and Raimondi, A. Simultaneous detection of membrane contact dynamics and associated Ca2+ signals by reversible chemogenetic reporters.
- Nat. Commun. 2024 – Bendel, A. M., Faure, A. J., Klein, D., Shimada, K., Lyautey, R., Schiffelholz, N., … & Diss, G. The genetic architecture of protein interaction affinity and specificity.
- ChemBioChem 2024 – Barrios, A., Hansen, D.T., Tu, J., Bouck, A.W. and Mathis, C.L. Multipartite Fluorogenic Sensors for Monitoring Tyrosine Phosphatase Activity.
- RSC Chem. Biol. 2024 – Torrey, Z.R., Halbers, L.P., Scipioni, L., Tedeschi, G., Digman, M.A. and Prescher, J.A. A versatile bioluminescent probe with tunable color.
- Microb. Cell Fact. 2024 – Jeon, E., Seong M., Hee S., & Ki J. Design of fully synthetic signal peptide library and its use for enhanced secretory production of recombinant proteins in Corynebacterium glutamicum.
- Microb. Cell Fact. 2024 – Mook, A., Herzog, J., Walther, P., Dürre, P., & Bengelsdorf, F. R. Lactate-mediated mixotrophic co-cultivation of Clostridium drakei and recombinant Acetobacterium woodii for autotrophic production of volatile fatty acids.
- Nat. Commun. 2024 – Wasserman, J. S., Faezov, B., Patel, K. R., Kurimchak, A. M., Palacio, S. M., Glass, D. J., … & Graña, X. FAM122A ensures cell cycle interphase progression and checkpoint control by inhibiting B55α/PP2A through helical motifs.
- J. Cell. Biol. 2024 – Li, X., Gamuyao, R., Wu, M.L., Cho, W.J., Kurtz, N.B., King, S.V., Petersen, R.A., Stabley, D.R., Lindow, C., Climer, L. and Shirinifard, A. A fluorogenic complementation tool kit for interrogating lipid droplet-organelle interaction.
- Nucleic Acids Res. 2024 – Lu, Z., Shen, Q., Bandari, N. C., Evans, S., McDonnell, L., Liu, L., … & Peng, B. LowTempGAL: a highly responsive low temperature-inducible GAL system in Saccharomyces cerevisiae.
- Comm. Biol. 2024 – Bogdanova, Y. A., Solovyev, I. D., Baleeva, N. S., Myasnyanko, I. N., Gorshkova, A. A., Gorbachev, D. A., … & Baranov, M. S. Fluorescence lifetime multiplexing with fluorogen activating protein FAST variants.
- bioRxiv 2024 – Verpoort, B., Amado, L., Vandensteen, J., Leysen, E., Dascenco, D., Vandenbempt, J., Lemmens, I., Wauman, J., Vennekens, K., Escamilla-Ayala, A. and Freitas, A.C.N. Cell-surface receptor-mediated regulation of synaptic organelle distribution controls dendritic spine maturation.
- bioRxiv 2024 – McKellar, J., García de Gracia, F., Aubé, C., Chaves Valadão, A.L., Tauziet, M., Arnaud-Arnould, M., Rebendenne, A., Neyret, A., Labaronne, E., Ricci, E. and Delaval, B. Human MX1 orchestrates the cytoplasmic sequestration of neo-synthesized influenza A virus vRNPs.
- Int. J. Mol. Sci. 2024 – Baleeva, N. S., Bogdanova, Y. A., Goncharuk, M. V., Sokolov, A. I., Myasnyanko, I. N., Kublitski, V. S., … & Baranov, M. S. A Combination of Library Screening and Rational Mutagenesis Expands the Available Color Palette of the Smallest Fluorogen-Activating Protein Tag nanoFAST.
- PLOS Pathog. 2024 – Anjou, C., Lotoux, A., Zhukova, A., Royer, M., Caulat, L. C., Capuzzo, E., … & Martin-Verstraete, I. The multiplicity of thioredoxin systems meets the specific lifestyles of Clostridia.
- Microb. Cell Fact. 2024 – Flaiz, M., Poehlein, A., Wiebke, W., Mook, A., Daniel, R., Dürre, P., & Bengelsdorf, F. Refining and illuminating acetogenic Eubacterium strains for reclassification and metabolic engineering.
- ACS Synth. Biol. 2024 – Peng, B., Weintraub, S. J., Lu, Z., Evans, S., Shen, Q., McDonnell, L., … & Vickers, C. E. Integration of Yeast Episomal/Integrative Plasmid Causes Genotypic and Phenotypic Diversity and Improved Sesquiterpene Production in Metabolically Engineered Saccharomyces cerevisiae.
- Russ. J. Bioorg. Chem. 2023 – Bogdanova, Y. A., Mineev, K. S., Baleeva, N. S., & Baranov, M. S. Effeсts of the 70th Amino Acid Residue on the Photostability of FAST Complexes.
- Front. Microbiol. 2023 – Tchagang, C. F., Mah, T. F., & Campbell-Valois, F. X. Anaerobic fluorescent reporters for live imaging of Pseudomonas aeruginosa.
- Front. Bioeng. Biotechnol. 2023 – Hocq, R., Bottone, S., Gautier, A., & Pflügl, S. A fluorescent reporter system for anaerobic thermophiles.
- bioRxiv 2023 – Anderson, D. M., Logan, M. G., Patty, S. S., Kendall, A. J., Borland, C. Z., Pfeifer, C. S., … & Merritt, J. L. Microbiome imaging goes à la carte: Incorporating click chemistry into the fluorescence-activating and absorption-shifting tag (FAST) imaging platform.
- Mater. Today Bio 2022 – Cao, Z., Wang, L., Liu, R., Lin, S., Wu, F., & Liu, J. Encoding with a fluorescence-activating and absorption-shifting tag generates living bacterial probes for mammalian microbiota imaging.
- J. Bacteriol. 2022 – Hernandez, E., & Costa, K. C. The fluorescence-activating and absorption-shifting tag (FAST) enables live-cell fluorescence imaging of Methanococcus maripaludis.
- Appl. Microbiol. Biotechnol. 2022 – Mook, A., Beck, M., Baker, J., Minton, N., Dürre, P., Bengelsdorf, F. Autotrophic lactate production from H2 + CO2 using recombinant and fluorescent FAST-tagged Acetobacterium woodii strains.
- ACS Synth. Biol. 2022 – Flaiz, M., Baur, T., Gaibler, J., Kröly, C., & Dürre, P. Establishment of Green- and Red-Fluorescent Reporter Proteins Based on the Fluorescence-Activating and Absorption-Shifting Tag for Use in Acetogenic and Solventogenic Anaerobes.
- Curr. Opin. Biotechnol. 2021 – Streett, H., Charubin, K., & Papoutsakis, E. T. Anaerobic fluorescent reporters for cell identification, microbial cell biology and high-throughput screening of microbiota and genomic libraries.
- Chem. Eur. J. 2021 – Chen, C., Tachibana, S. R., Baleeva, N. S., Myasnyanko, I. N., Bogdanov, A. M., Gavrikov, A. S., … & Fang, C. Developing Bright GFP‐Like Fluorogens for Live‐Cell Imaging with Nonpolar Protein‐Chromophore Interactions.
- npj Biofilms Microbiomes 2021 – Monmeyran, A., Benyoussef, W., Thomen, P., Dahmane, N., Baliarda, A., Jules, M., … & Henry, N. Four species of bacteria deterministically form a stable biofilm in a millifluidic channel: assembly principles.
- Russ. J. Bioorg. Chem. 2021 – Sokolov, A. I., Myasnyanko, I. N., Baleeva, N. S., & Baranov, M. S. Styrene Derivatives of Indole and Pyranone as Fluorogenic Substrates for FAST Protein.
- Nat. Commun. 2021 – Lu, Z., Peng, B., Ebert, B. E., Dumsday, G., & Vickers, C. E. Auxin-mediated protein depletion for metabolic engineering in terpene-producing yeast.
- Biotechnol. Biofuels 2021 – Flaiz, M., Ludwig, G., Bengelsdorf, F. R., & Dürre, P. Production of the Biocommodities Butanol and Acetone from Methanol with Fluorescent FAST-tagged Proteins using Metabolically Engineered Strains of Eubacterium Limosum.
- Chem. Sci. 2021 – Mineev, K. S., Goncharuk, S. A., Goncharuk, M. V., Povarova, N. V., Baleeva, N. S., Smirnov, A. Y., … & Baranov, M. S. NanoFAST: Structure-based design of a small fluorogen-activating protein with only 98 amino acids.
- Chem. Eur. J. 2020 – Myasnyanko, I. N., Gavrikov, A. S., Zaitseva, S. O., Smirnov, A. Y., Zaitseva, E. R., Sokolov, A. I., … & Baranov, M. S. Color tuning of fluorogens for FAST fluorogen‐activating protein.
- Sci. Rep. 2020 – Chekli, Y., Peron-Cane, C., Dell’Arciprete, D., Allemand, J. F., Li, C., Ghigo, J. M., … & Beloin, C. Visualizing the dynamics of exported bacterial proteins with the chemogenetic fluorescent reporter FAST.
- PLoS Pathog. 2020 – Peron-Cane, C., Fernandez, J. C., Leblanc, J., Wingertsmann, L., Gautier, A., Desprat, N., & Lebreton, A. Fluorescent secreted bacterial effectors reveal active intravacuolar proliferation of Listeria monocytogenes in epithelial cells.
- Mbio 2020 – Charubin, K., Modla, S., Caplan, J. L., & Papoutsakis, E. T. Interspecies Microbial Fusion and Large-Scale Exchange of Cytoplasmic Proteins and RNA in a Syntrophic Clostridium Coculture.
- Appl. Environ. Microbiol. 2020 – Charubin, K., Streett, H., & Papoutsakis, E. T. Development of Strong Anaerobic Fluorescent Reporters for Clostridium acetobutylicum and Clostridium ljungdahlii Using HaloTag and SNAP-tag Proteins.
- Appl. Environ. Microbiol. 2019 – Streett, H. E., Kalis, K. M., & Papoutsakis, E. T. A strongly fluorescing anaerobic reporter and protein-tagging system for Clostridium organisms based on the fluorescence-activating and absorption-shifting tag protein (FAST).
- Chem. Eur. J. 2019 – Povarova, N. V., Zaitseva, S. O., Baleeva, N. S., Smirnov, A. Y., Myasnyanko, I. N., Zagudaylova, M. B., … & Mishin, A. S. Red‐shifted substrates for FAST fluorogen‐activating protein based on the GFP‐like chromophores.
- ACS Chem. Biol. 2019 – Smith, E. M., Gautier, A., & Puchner, E. M. Single-molecule localization microscopy with the fluorescence-activating and absorption-shifting tag (FAST) system.
- Nanoscale 2019 – Venkatachalapathy, M., Belapurkar, V., Jose, M., Gautier, A., & Nair, D. Live cell super resolution imaging by radial fluctuations using fluorogen binding tags.
- Metab. Eng. 2018 – Charubin, K., Bennett, R. K., Fast, A. G., & Papoutsakis, E. T. Engineering Clostridium organisms as microbial cell-factories: challenges & opportunities.
- Sci. Rep. 2018 – Monmeyran, A., Thomen, P., Jonquière, H., Sureau, F., Li, C., Plamont, M. A., … & Henry, N. The inducible chemical-genetic fluorescent marker FAST outperforms classical fluorescent proteins in the quantitative reporting of bacterial biofilm dynamics.
- Nat. Methods 2017 – Emanuel, G., Moffitt, J. R., & Zhuang, X. High-throughput, image-based screening of pooled genetic-variant libraries.